Data related to this publication
by Wadim Matochko, Cory S. Li, Sindy K.Y. Tang, Ratmir Derda*, Nucl. Acids Res. (2013) doi: 10.1093/nar/gkt1104
To download raw files, please visit this site:
http://www.chem.ualberta.ca/~derda/parasitepaper/rawfiles/
Please use this link to download the updated ZIP file that contains all MatLab Scipts from this paper
MatLab Scripts Patched on Feb 5, 2014 (patched script can run without MatLab BioInformatic Toolbox)
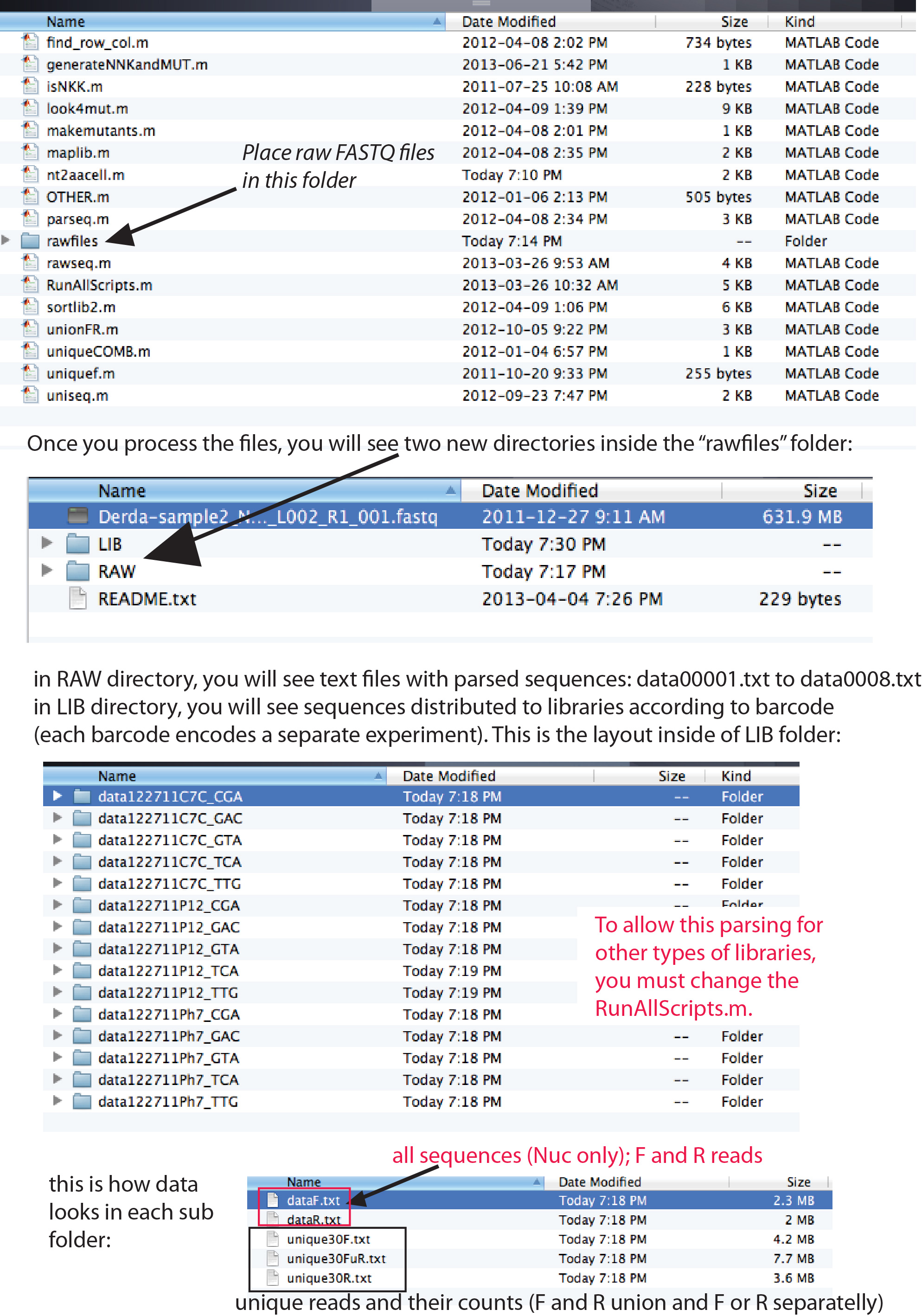
Once you unzip the folder, follow this simple instruction and use a sameple FASTQ file from here (caution, it is 603 Mb!!) : http://www.chem.ualberta.ca/~derda/parasitepaper/Derda-sample2_NoIndex_L002_R1_001.fastq
Disclaimer: the scripts in MatLab folder are designed for processing of only Ph.D. libraries described in this paper amplified using only primers described in this paper. The script will not work for any libraries or any primers. We note that scripts can be easily modified to fit any phage library of similar length (knowledge of MatLab is recommended). To attempt a modification on your own, please see some guidance here. For assistance in modification please contact Ratmir Derda (coresponding author).